| Suggestion: tautomer-check [message #886] |
Wed, 06 May 2020 15:40  |
 nbehrnd
nbehrnd
Messages: 240
Registered: June 2019
|
Senior Member |
|
|
DataWarrior's model to assign Druglikeness depends on the encoded structure a
tautomer is represented. With Ambit-Tautomer, Kochev et al. published an open
source tool (Java based), to predict tautormers and to rank their likelyhood.
If wanted, their executable may be run without the larger Ambit framework, just
by
java -jar ambit-tautomers-2.0.0-SNAPSHOT.jar
Thus, I would like to suggest DataWarrior could implement a function to check
if the structures to consider could reasonably yield a tautomer worth to probe
equally. This equally could be complementary to your recently published reference
tautomer.dwar.
Out of curiosity, I drew a pyridone, a pyrazole, and a thalomide with ACD ChemSketch
in two tautomeric forms, exported the SMILES strings (as defined by ACD ChemSketch)
into a .smi file:
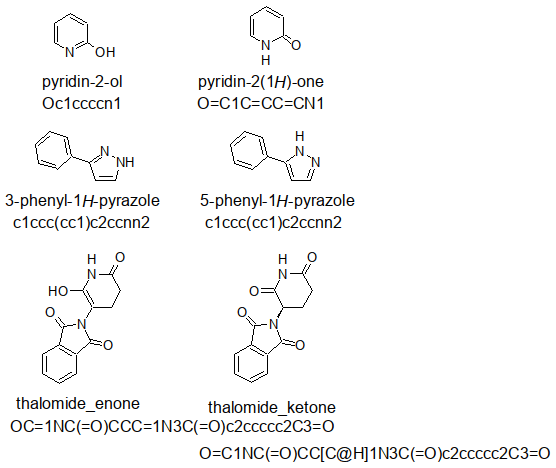
With openbabel, it was converted into a .sdf accessible for DataWarrior by
obabel -ismi tautomers.smi -osd -O tautomers.sdf
successfully read and used to compute the Druglikeness. At least as the examples
about «hydroxypyridine» and the enol form of the thalomide differ in the results
examined.
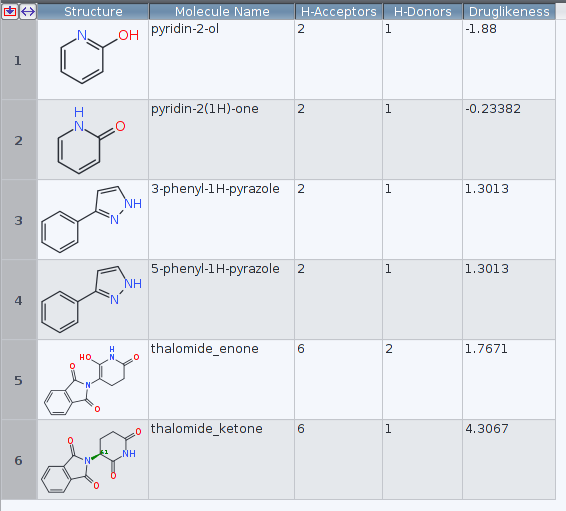
To ease replication of the findings, the relevant files are provided below.
Publication about Ambit-Tautomer: https://doi.org/10.1002/minf.201200133
github-entry about Ambit-Tautomer: https://github.com/ideaconsult/apps-ambit/tree/master/tautom ers-example
|
|
|
|
