| Importing compounds with assay data from Pubchem into Datawarrior [message #1354] |
Fri, 23 July 2021 13:12  |
Jo W
Messages: 34
Registered: July 2021
|
Member |
|
|
Many thanks Thomas and colleagues
Data warrior is a "life saver" for non coding people like me - thank you so much for you and your team for developing it.
Its also one of ( I think it is THE) easiest software to use for a variety of informatics / prediction modeling etc.
Importing / uploading to Datwarrior is usually really straightforward compared to other programs.
For example, it's possible to download compounds from Pubchem and via a text / csv file import into Datawarrior with no editing. This works really well and is easy to do.
However when trying to download, say 30,000 compounds that are also P450 inhibitors an error comes up, maybe because the request has too many data points or maybe it cant be done via a web page (Pubchem talks about Restful / ftp but for non coding people this seems very difficult to do - 2 hours trying to figure this out did not prove fruitful!
My questions/requests/comments are:
1. A request - Can Datawarrior create a built in search for Pubchem along similar lines to the Chembl facility? Pubchem seems to have so many more compounds with accompanying biodata in it compared to Chembl.
2. In the meantime - how can you import compounds with specific biodata from Pubchem into datawarrior?
Can you use Datawarriors URL import? I tried this for example with a list of P450 enzyme inhibition active compounds from Pubchem (by copying and pasting the url, but this did not work:
https://pubchem.ncbi.nlm.nih.gov/protein/P10633#section=Chem icals-and-Bioactivities
Any ideas regarding the URL or any other "non coding" ways to get data form Pubchem into Datawarrior? As stated above, you cannot simply download a csv file from Pubchem (at least for obtaining biodata) from the above link as an error occurs, even though Pubchem provides a downlaod "button" for it.
Many thanks in advance
Jon
[Updated on: Fri, 23 July 2021 13:15] Report message to a moderator |
|
|
|
| Re: Importing compounds with assay data from Pubchem into Datawarrior [message #1355 is a reply to message #1354] |
Fri, 23 July 2021 15:17   |
 thomas
thomas
Messages: 684
Registered: June 2014
|
Senior Member |
|
|
Hi Jon,
unfortunately, I am afraid, I cannot help you much. I never fully explored what is needed to keep a fully updated copy of PubChem's bioactivity on a structure searchable server, but it always seemed to me that it would be beyond the effort, I would be willing to invest. I just tried to download a few data files from the PubChem website (pubchem.ncbi.nlm.nih.gov). Some download button scripts simply didn't do anything (Firefox on Ubuntu), but with most of them I could download either a CSV, SDF or gzipped SDF. All of those I could open successfully in DataWarrior ('.sdf.gz' need the newest dev update). Unless somebody else would volunteer to write the code to retrieve the PubChem bioassay data and merge it with the structures where needed and keep everything updated on a searchable server engine, I won't have the time to provide easy DataWarrior search access.
If you manage to download PubChem data files (csv or sdf) and have trouble reading them into DataWarrior because of limited csv/sdf capabilities, then please let me know. I would do my best to fix these issues on the DataWarrior side.
Thomas
|
|
|
|
|
|
| Re: Importing compounds with assay data from Pubchem into Datawarrior [message #1358 is a reply to message #1357] |
Sun, 25 July 2021 17:18   |
Jo W
Messages: 34
Registered: July 2021
|
Member |
|
|
Apologies for the "winking smileys" - I did not want to include them in my posts - they just "appeared" when I used "underline".
Also, I have noticed 43 people have looked at this thread in the space of 24 hours. Please folks add some help or some suggestions here. I can't believe I am the only one wanting to add / upload PubChem data to Datawarrior, this fantastic piece of software.
Or PM me if you prefer with any helpful tips or suggestions.
Many thanks in advance
[Updated on: Sun, 25 July 2021 17:18] Report message to a moderator |
|
|
|
| Re: Importing compounds with assay data from Pubchem into Datawarrior [message #1360 is a reply to message #1358] |
Mon, 26 July 2021 21:55   |
 nbehrnd
nbehrnd
Messages: 213
Registered: June 2019
|
Senior Member |
|
|
Dear Jon,
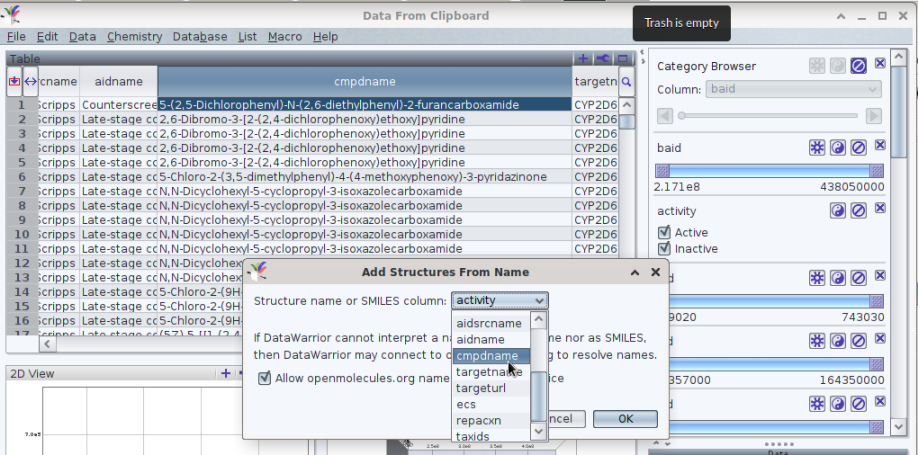
there was indeed an problem to access and subsequently keep a local copy of the .csv files. By now, yhe support by PubChem was able to correct the underlying error. The downloaded file contains a (late) column labelled «cmpdname»
Open the file in a text editor to copy all content into the working memory of the computer. In DataWarrior, paste these information via Edit -> Paste Special -> New From Data With Header Row. Establish the structures via Chemistry -> Add Structures From Name which opens a new interface. To guide DW's action, select the column of interest, «cmpdname».
Norwid
[Updated on: Tue, 27 July 2021 23:19] Report message to a moderator |
|
|
|
|
|
|
|

 Search
Search Help
Help Members
Members Register
Register Login
Login Home
Home










