|
|
|
|
|
|
|
|
| Re: Pentavalent carbonyl issue (urea formation?) [message #1942 is a reply to message #1941] |
Sat, 24 June 2023 16:53   |
 thomas
thomas
Messages: 741
Registered: June 2014
|
Senior Member |
|
|
Thank you for the files. To me it seems that your atom mapping is incorrect. The reactants fragments consisting out of a carbonyl group with attached bridge bond to nitrogen should not break bonding in the reaction, thus they should appear in the product the same way with the same mapping numbers. I have attached the updated macro file, which seems to work fine now.
The upper reaction shows your original mapping, the lower one shows the updated mapping:
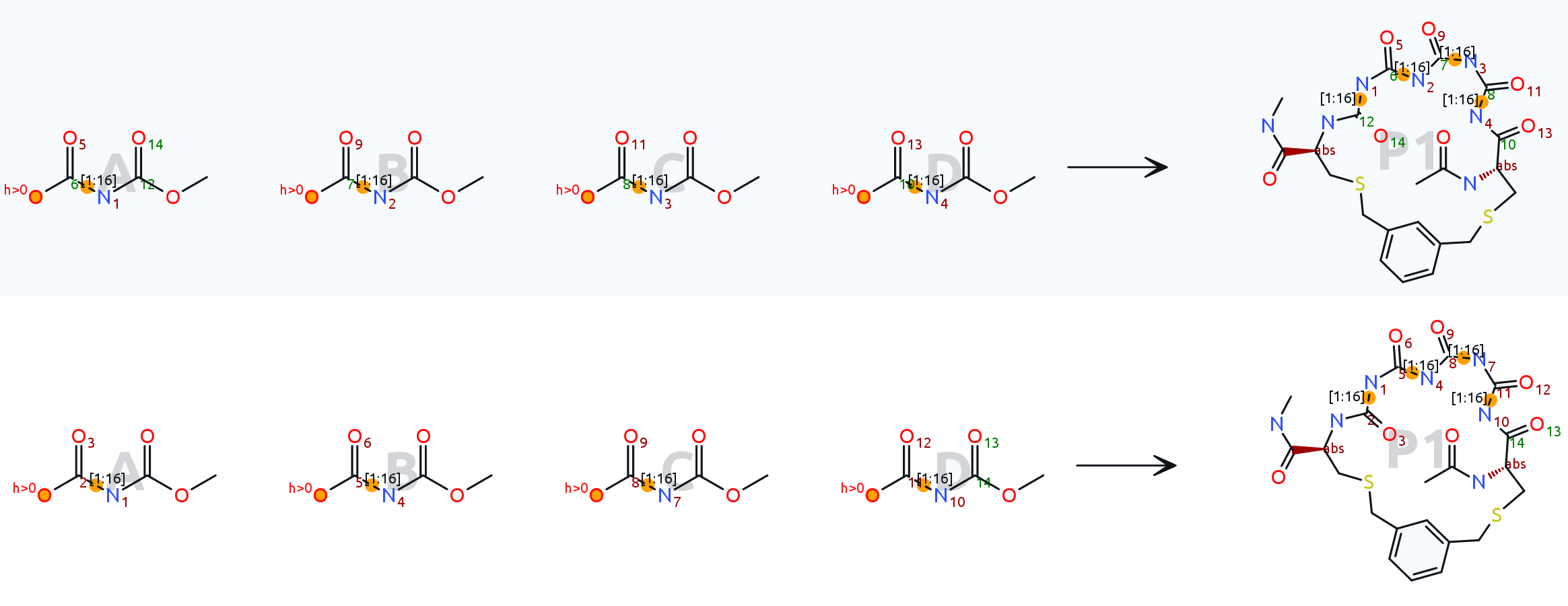
[Updated on: Sat, 24 June 2023 16:54] Report message to a moderator |
|
|
|
|
|
| Re: Pentavalent carbonyl issue (urea formation?) [message #1944 is a reply to message #1943] |
Sun, 25 June 2023 13:31   |
 thomas
thomas
Messages: 741
Registered: June 2014
|
Senior Member |
|
|
If I import and run the macro from my last message, then I get the following product (with both versions 5.5.0 and with the current dev update):
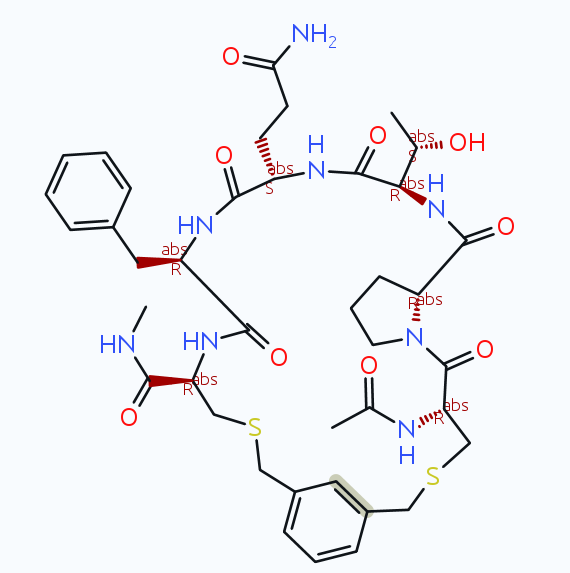
If you use the same version on MacOS, Linux, or Windows, then it should behave exactly the same. Which version do you use anyway? The official 5.5.0, that mean the most recent (but 2 years old) official installer? Then I would recommend to update to the newest dev build (on the download page click on the 'read and understood checkbox, then click the link in the small print for you platform and download the respective archive with patch files for your installation)
Did you manually map as many atoms as it needs for auto-mapper to correctly map all remaining atoms of the reaction? If not, I suggest to remove all manual mapping and start from scratch to map at least all four O=C...N fragments manually.
Another tip: Typically, one does not use atom or bond query features on the product side of the generic reaction. Bridge bonds are an exception, because whatever matches on the reactant side needs to be reconstructed on the product side. If all your real reactants have the same bridge length, then I would remove the bridge bond from the reaction and replace it with real bonds and atoms.
One more tip: A part of the new medium sized ring comes from unknown components. For the sake of complete and perfect mapping and a stoichiometrically complete reaction you might introduce a fifth reactant with the missing atoms, that is always the same.
|
|
|
|
|
|
