| Aromaticity perception [message #967] |
Thu, 25 June 2020 22:38  |
 richards99
richards99
Messages: 42
Registered: May 2020
Location: UK
|
Member |
|
|
Hi,
There appears to be consistent issues with the import of certain aromatised structures.
Ones I especially notice are N-methylated pyridinones, pyrimidinones, and bicyclic structures.
Data Warrior puts double bonds into the wrong positions, quaternising nitrogens.
It would be useful if the aromaticity perception can be improved otherwise it creates lots of invalid smiles which require manually altering.
Thanks,
Simon.
|
|
|
|
|
|
|
|
|
|
| Re: Aromaticity perception [message #974 is a reply to message #971] |
Sun, 28 June 2020 23:57   |
 nbehrnd
nbehrnd
Messages: 240
Registered: June 2019
|
Senior Member |
|
|
Hi Simon,
it was possible to replicate the problem with the .sdf shared by you. Tentatively, the
problem is caused by the existence of multiple SMILES dialects by different programs which
may be an obstacle for DW. I thus recommend to pass the .sdf to openbabel to pass the files'
content into a new .sdf file to solve the issue. Here, DataWarrior (version 5.2.1, native
installation in Linux Debian) and openbabel (version 3.1.0, June 9, 2020) were used.
Enter the directory containing the .sdf in question. From the terminal (Linux, Mac) or
cmd.exe (Windows) provide a instruction in line of
obabel -isdf Aromatised.sdf -osdf -O Aromatised_passed_obabel.sdf
With only eight molecules, this is a quick operation. Comparing the original and the new /
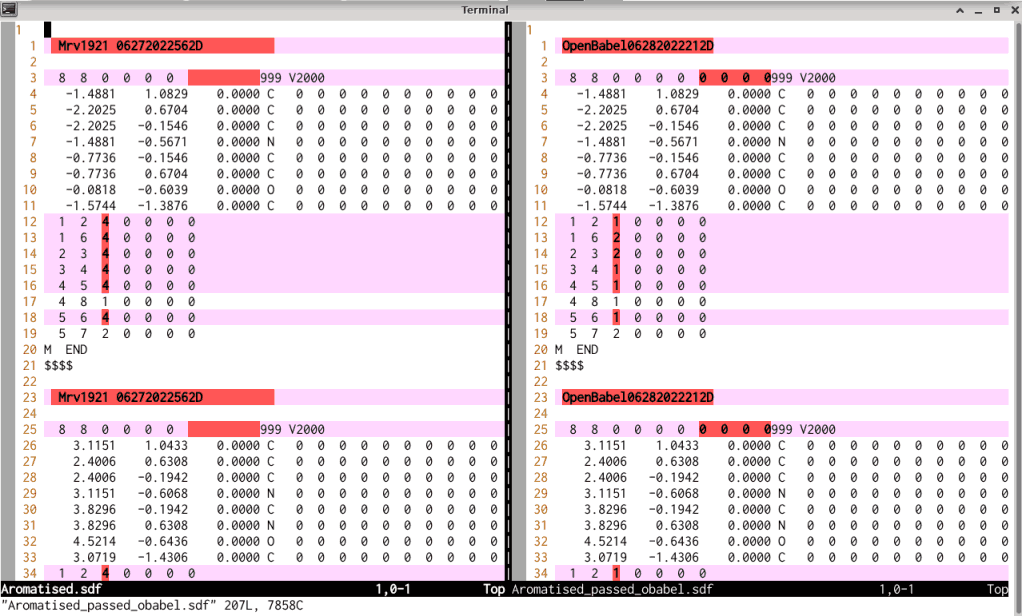
derived .sdf file with each other shows that the connection table in the files is adjusted,
as shown in the screen photo below:
More importantly, the issues with tetravalent nitrogen atoms are resolved:
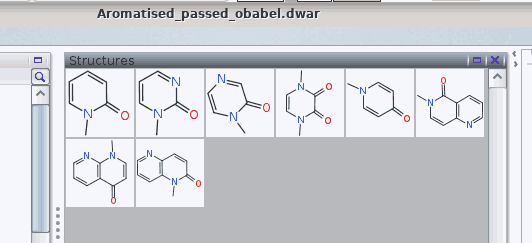
The .dwar eventually obtained is provided as an attachment of this answer.
Openbabel is a freely available program running on Windows, Mac, and Linux to interconvert
chemical formats. Its code is open on GitHub, which equally hosts the executables. The
documentation may be accessed online, or offline. If wanted, a GUI may provide you an easier
entry into a selection of its functions, too.
---
It is equally possible to convert the SMILES as provided into an .sdf, too. The command then
would be
obabel -ismi probe.smi -osdf -O probe.sdf
to lead to the same result as above, or copy-pasting (without header row) the SMILES directly
into DW. Both .smi and .sdf of this approach equally are provided here.
Norwid
https://github.com/openbabel/openbabel
https://github.com/openbabel/openbabel/releases/tag/openbabe l-3-1-1
https://open-babel.readthedocs.io/en/latest/
https://open-babel.readthedocs.io/_/downloads/en/latest/pdf/
[Updated on: Sun, 28 June 2020 23:58] Report message to a moderator |
|
|
|
| Re: Aromaticity perception [message #980 is a reply to message #974] |
Thu, 02 July 2020 14:41  |
 thomas
thomas
Messages: 742
Registered: June 2014
|
Senior Member |
|
|
many thanks Simon and Norwid for pointing to this issue suggesting work-arounds.
The problem was that DataWarrior didn't expect finding compounds with aromatic bond types in molfiles,
which are based on the Daylight aromaticity model into the bargain, e.g. having carbonyl carbon atoms being
maked as aromatic. This is unusual for two reasons: First, molfiles typically store alternating single
and double bonds for aromatic rings rather than using the delocalized bond type, unless it encodes
a substructure with query features. Second, for the rare cases that the delocalized bond types may be used
one would expect an MDL/Symex/Hueckel aromaticity concept to be applied.
Nevertheless, since Marvin Sketch seems to read SMILES based atom aromaticity encodings and writes them directly into
written molfiles using aromatic bond types, I have updated DataWarrior to normalize this kind of encoding before
generating and writing idcodes (DataWarrior's canonical structure representation) into its native files.
The current developments version should not have this issue anymore.
Thomas
|
|
|
|
