| Re: tautomer & protomer display [message #796 is a reply to message #793] |
Mon, 02 March 2020 18:34  |
 thomas
thomas
Messages: 742
Registered: June 2014
|
Senior Member |
|
|
DataWarrior stores chemical structures in memory and within its native files as very compact canonical string representation, so-called idcodes. SD-Files and SMILES are converted to idcodes, when they are read as input file (or from the clipboard). After reading the original, DataWarrior performs some structure normalization before creating the canonical idcode. One example is to unify the nitro group as -N(+)(=O)-O(-), which in some input SMILES or SD-files comes as -N(=O)=O. Part of the normalization is also to neutralize charged atoms as your example:
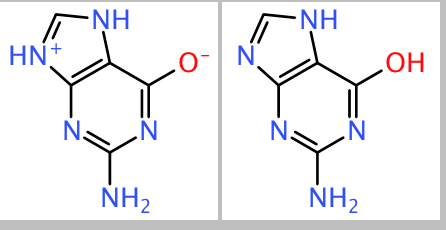
Because of these normalization rules DataWarrior considers these both structures to be equivalent and, thus, creates the same idcode for them, which actually represents the second form of the structure.
Note that you can manually change this structure again within DataWarrior by adding charges and implicitly adding/removing hydrogen atoms to come back to the first structure. If you save the file and reopen, the bipolar structure is retained. The idea behind that is that structure files from different sources may use different conventions for representing the same thing. DataWarrior does its best to normalize. When a chemist, however, directly within DataWarrior edits a structure, then he probably means exactly what he is drawing.
-
 Attachment: t.png
Attachment: t.png
(Size: 16.56KB, Downloaded 942 times)
[Updated on: Mon, 02 March 2020 18:36] Report message to a moderator |
|
|
|
