| Generating derivatives of parent structure [message #2027] |
Sun, 05 November 2023 10:09  |
 lutek14
lutek14
Messages: 3
Registered: October 2019
|
Junior Member |
|
|
Hello,
I appreciate the quality of the program, but require some assistance. I have an original structure and wish to generate derivatives by substituting specific locations by certain residues. Is this achievable? Alternatively, could you recommend any freely available software that can accomplish this task efficiently?
Thanks
|
|
|
|
| Re: Generating derivatives of parent structure [message #2028 is a reply to message #2027] |
Sun, 05 November 2023 19:14   |
 nbehrnd
nbehrnd
Messages: 240
Registered: June 2019
|
Senior Member |
|
|
Hello,
this taps into the generation of a combinatorial library you find below Chemistry -> enumerate combinatorial library. Either you find your reaction among the templates DW ships by default, or you create your own reaction pattern. Tab «reactants» allows you to use molecules already defined in a .dwar, or .sdf; or to let DW select ones which are commercially available (criteria are adjustable).
See chapter «chemical structures», section «combinatorial library» in DW's help or the online web page[1] for additional information. It however is time well invested to follow Isabelle Girault's video tutorial about the topic «RSC CICAG Open Source Tools for Chemistry Workshops:- Advanced DataWarrior»[2] recorded in 2021 around 47:18 min:s on youtube. (There is sequal «DataWarrior workshop by Isabelle Giraud»[3] by her to complement the insight about DW, too.)
Regards,
Norwid
[1] https://openmolecules.org/help/chemistry.html#VirtualLibrari es
[2] https://www.youtube.com/watch?v=mQCf9GakQW0
[3] https://www.youtube.com/watch?v=Is2hLqqSFvM
[Updated on: Sun, 05 November 2023 19:15] Report message to a moderator |
|
|
|
|
|
|
|
| Re: Generating derivatives of parent structure [message #2048 is a reply to message #2029] |
Sat, 09 December 2023 00:42  |
 thomas
thomas
Messages: 742
Registered: June 2014
|
Senior Member |
|
|
You could also use the "Build Evolutionary Library..." functionality. This creates new molecules from a starting generation by random small modifications. You can also define parts of the molecule that shall not be modifed by just selecting it. I tried that with your molecule and selected all but 4 ring positions. This causes random modifications at all selected atoms, which includes ring cleavages.
Then you are supposed to add fitness criteria, which are used to value the quality of created structures. Those structures, that match your criteria best will be kept and form at the same time the parent generation for the next round of random changes. I just defined a simple criterion: the molecular weight shall be not higher than 240. Otherwise I just use default options for everything else. The settings look like this:
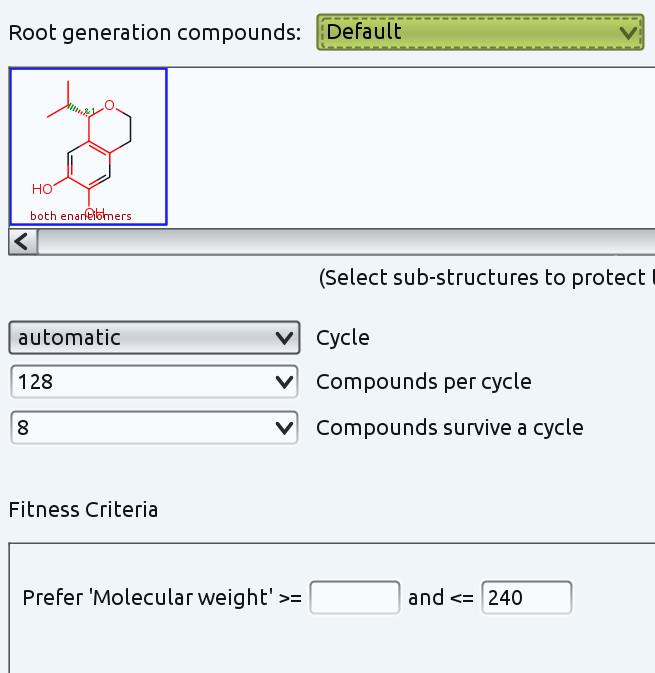
In less than one second DataWarrior created 267 different molecules in 15 generations that all contain the selected (red) part of your defined starting structure, with lots of variations regarding substitution, ring size, partially open ring structures and molecular weights always below 240. The picture shows a small subset.
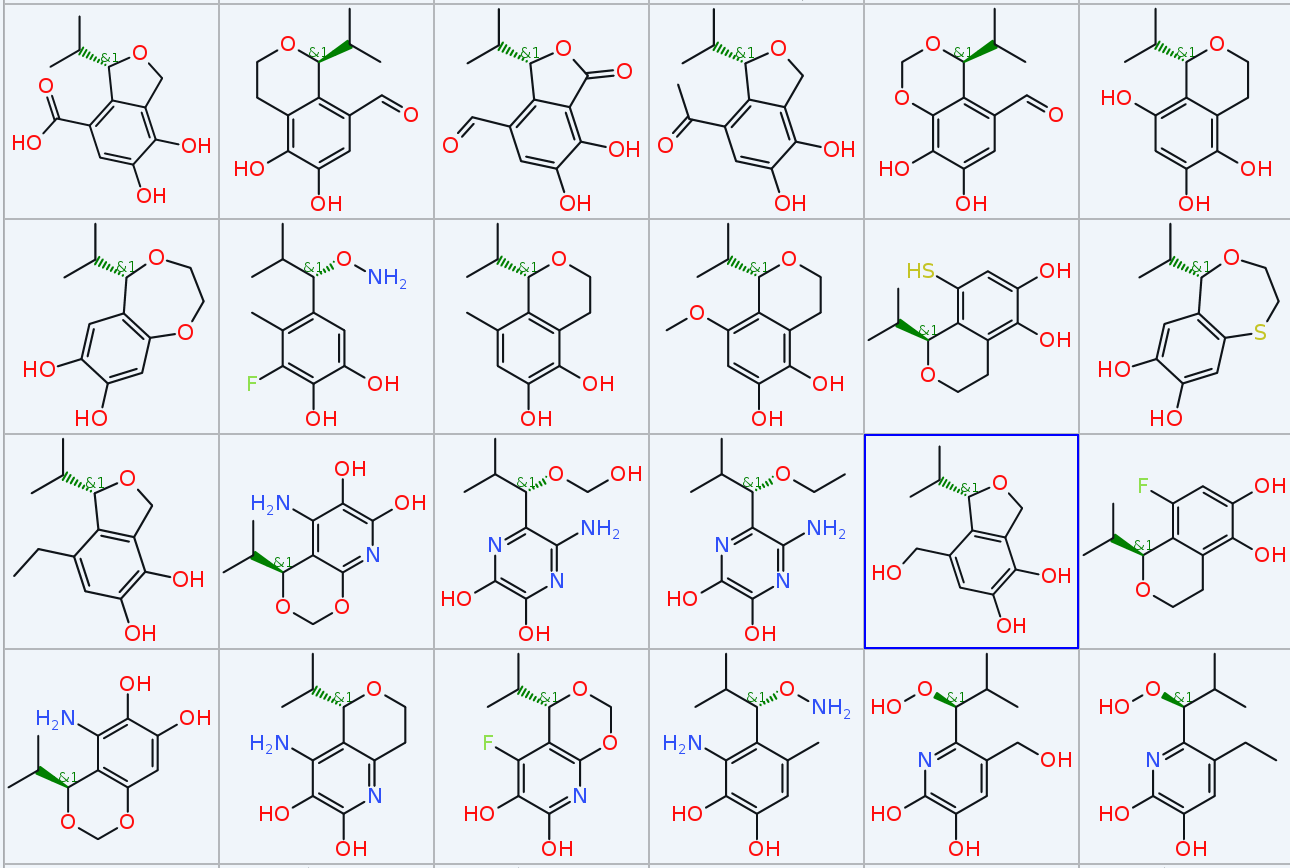
Of course, you can be more specific with fitness criteria, e.g. use chemical similarities, pharmacophore similaries or even a docking score...
-
 Attachment: temp2.png
Attachment: temp2.png
(Size: 139.27KB, Downloaded 698 times)
-
 Attachment: temp1.png
Attachment: temp1.png
(Size: 40.66KB, Downloaded 749 times)
[Updated on: Sat, 09 December 2023 00:46] Report message to a moderator |
|
|
|
