| HBD/HBA [message #1588] |
Fri, 08 April 2022 15:13  |
 chemsandra
chemsandra
Messages: 1
Registered: April 2022
|
Junior Member |
|
|
I have a question about hydrogen bond donors (HBD) and hydrogen bond acceptors (HBA). I was wondering how DataWarrior software defines HBD and HBA. Is there any literature reference about this?
I used KNIME and DataWarrior to calculate the number of HBA and HBA. However, the result of using both software is different.
I would be grateful for the help with this.
|
|
|
|
|
|
| Re: HBD/HBA [message #1863 is a reply to message #1608] |
Mon, 06 March 2023 20:59   |
P_Fitz
Messages: 17
Registered: November 2019
|
Junior Member |
|
|
|
Is this an accurate way to do HBA/HBD? I wouldn't consider an amide -N an H bond acceptor.
|
|
|
|
| Re: HBD/HBA [message #1865 is a reply to message #1863] |
Mon, 06 March 2023 23:28   |
 nbehrnd
nbehrnd
Messages: 240
Registered: June 2019
|
Senior Member |
|
|
Initially for curiosity, I submitted a few simple molecules to the assignment to H-bond donors, and acceptors
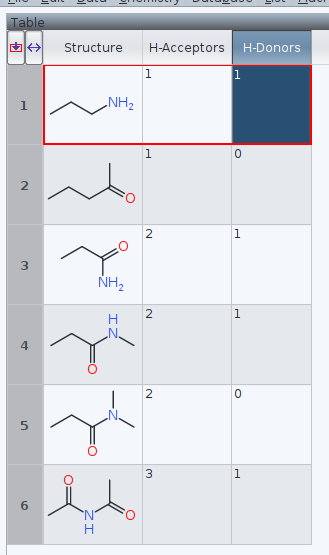
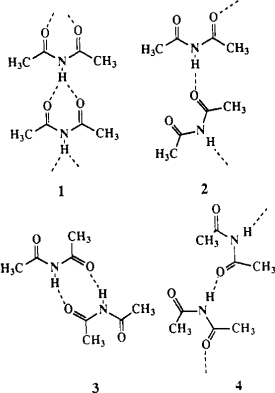
DW assigns last entry 6, diacetamide, 3 acceptors and 1 donor. This contrasts to Etter's paper[1] with two accptors and one donor only.
Do follow medicinal chemistry and solid state chemistry/crystallography different conventions here?
With regards,
Norwid
[1] Etter, M. C. Encoding and decoding hydrogen-bond patterns of organic compounds. Acc. Chem. Res. 1990, 23, 120-126 (DOI https://doi.org/10.1021/ar00172a005).
[2] Etter, M. C.; MacDonald, J. C.; Bernstein, J. Graph Set Analysis of Hydrogen-Bond Patterns in Organic Crystals. Acta Cryst. B 1990, 46, 256-262 (DOI
https://doi.org/10.1107/S0108768189012929)
|
|
|
|
| Re: HBD/HBA [message #1866 is a reply to message #1865] |
Tue, 07 March 2023 09:25   |
 zhentg
zhentg
Messages: 32
Registered: March 2020
|
Member |
|
|
Great that this topic is discussed.
We also found the HBA/HBD counts are different from those provided by PubChem(calculated by CACTVS).
Interestingly, RDkit HBA/HBD counts seems to be similar to DW.
It will be great if the CACTVS HBA/HBD count algrithm be adapted in DW.
Datawarrior is a good tool
|
|
|
|
| Re: HBD/HBA [message #1874 is a reply to message #1866] |
Sat, 18 March 2023 17:25  |
 thomas
thomas
Messages: 742
Registered: June 2014
|
Senior Member |
|
|
|
Admittedly, DataWarrior's (or RDkit's) simple approach of calculating HBA/HBD do not really reflect the reality in some cases. They just use the method applied in Lipinsky's 'rule of five' paper. This way DataWarrior's calculated HBA/HBD values can directly be used in the 'rule of five' context.
|
|
|
|
