| Re: Macro for evolutionary library (EL) in Datwarrior (DW) [message #1703 is a reply to message #1702] |
Tue, 23 August 2022 21:58   |
 nbehrnd
nbehrnd
Messages: 240
Registered: June 2019
|
Senior Member |
|
|
Dear Jon,
while converting the macro you reported as text into a .dwam file (for convenience of further use attached below), I noticed multiple pairs where «a seed» reporting a molecules' structure in idcodes (the line starts with "startSet=") is followed by an other line beginning with "paramConfig0". Assuming the first line counted as line "1", in the present .dwam this pattern occurs thrice, i.e. for lines (6, 9), (29, 32), and (60, 63).
Simultaneously, I read on the second line of these a high SkelSphere threshold, 100, e.g.
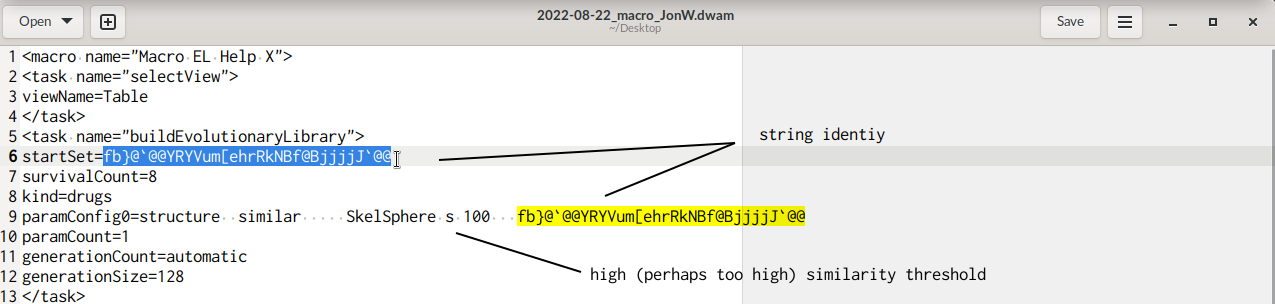
My speculation would be that this requested similarity might be too strict to yield multiple (new) molecules, perhaps especially because (based on your screen photos submitted) there only is one molecule to define the fitness of molecules to retain for a report / subsequent use.
Norwid
|
|
|
|
