| Re: Macro for evolutionary library (EL) in Datwarrior (DW) [message #1697 is a reply to message #1696] |
Tue, 09 August 2022 22:06   |
 nbehrnd
nbehrnd
Messages: 240
Registered: June 2019
|
Senior Member |
|
|
Dear Jon,
when generating an evolutionary library, DataWarrior offers (at least) twice to use a set of molecules from a .sdf or a .dwar as reference; in the screen photo below, they are marked by (A) and (B).
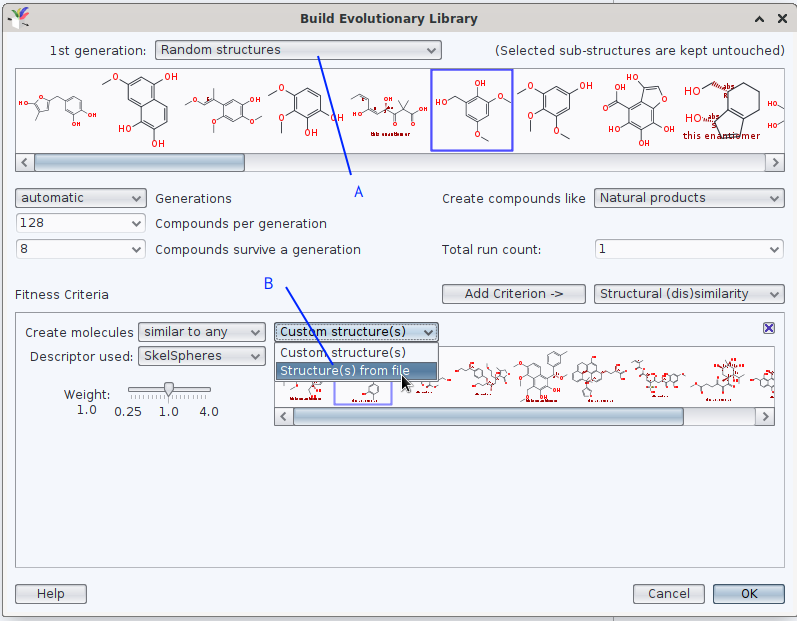
Should your question be understood in lines of «instead of molecules of an (of one) EL to be similar enough to a set of e.g., 20 molecules defined in box A, you would rather generate 20 EL where each of these reference molecules is a unique seed for an individual EL (more like an iteration)»?
Anyway, a .dwar and macro .dwam to represent a minimal working example (even if it currently stops after the first molecule of reference) to complement your question would be welcomed.
Norwid
[Updated on: Tue, 09 August 2022 22:19] Report message to a moderator |
|
|
|
