|
|
| Re: Enumeration of Libraries [message #1502 is a reply to message #1495] |
Thu, 10 February 2022 11:04  |
 thomas
thomas
Messages: 742
Registered: June 2014
|
Senior Member |
|
|
DataWarrior's concept behind the combinatorial enumeration is the definition of substructure based generic reaction. The reactant substructures are supposed to be substructures of real building blocks. That means they should contains the functional groups which are needed for the reaction to work. Atom and bond query features as well as exclude groups can be used as constraints, such that a substructure search in real building blocks only matches those compounds, which are reactive enough for the reaction to work.
Different from this concept combinatorial libraries are sometimes defined using pre-processed building blocks, where a real chemical functionality is replaced by pseudo atoms (U,Np,...), which basically represent a connection point. Two or more lists of pre-processed (sometimes called 'clipped reactants') can be joined by removing matching pseudo atoms and connecting their neighbours by a new single bond. This concept has its limitations, but is fast to do once the clipped libraries exist. While DataWarrior's reactor was not built with this concept in mind, it can be used (with limitations) for both, pre-processing building blocks to build a clipped library and to process multiple clipped libraries to generate product structures. The limitation I see is that a clipped library that contains multiple connection points (pseudo atoms) must not contain building blocks of type U-X-Np and U-X---Y-Np at the same time. You can define the generic reaction as follows.
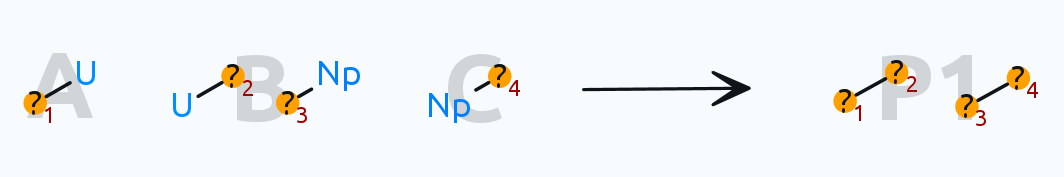
If some of your B reactants have U and Np connected to the same atom, then these are missed by the reaction above. For those you may do a second run with this reaction and afterwards merge the compounds generated by both runs.
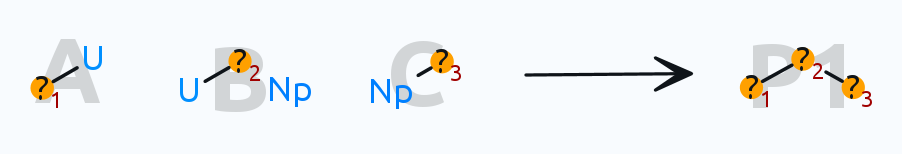
(Note: there is an invisible single bond in the second reactant between ? and Np)
Hope this answers the question...
Thomas
-
 Attachment: t.png
Attachment: t.png
(Size: 18.06KB, Downloaded 954 times)
-
 Attachment: t2.png
Attachment: t2.png
(Size: 16.03KB, Downloaded 894 times)
[Updated on: Thu, 10 February 2022 11:06] Report message to a moderator |
|
|
|
