Home » DataWarrior » Bug Reports » Stereoisomers conformer generation and Inchikeys
| Stereoisomers conformer generation and Inchikeys [message #1388] |
Tue, 31 August 2021 17:08  |
 mattiafelice.palermo
mattiafelice.palermo
Messages: 3
Registered: June 2021
|
Junior Member |
|
|
Dear Thomas,
I'm not 100% sure this is a bug as I am not particularly educated about InChIKeys generation and I am a bit rusty regarding stereochemistry.
I have calculated the lowest energy conformers for a molecule with two cis/trans stereocenters (limiting to 1 conformer per stereoisomer):
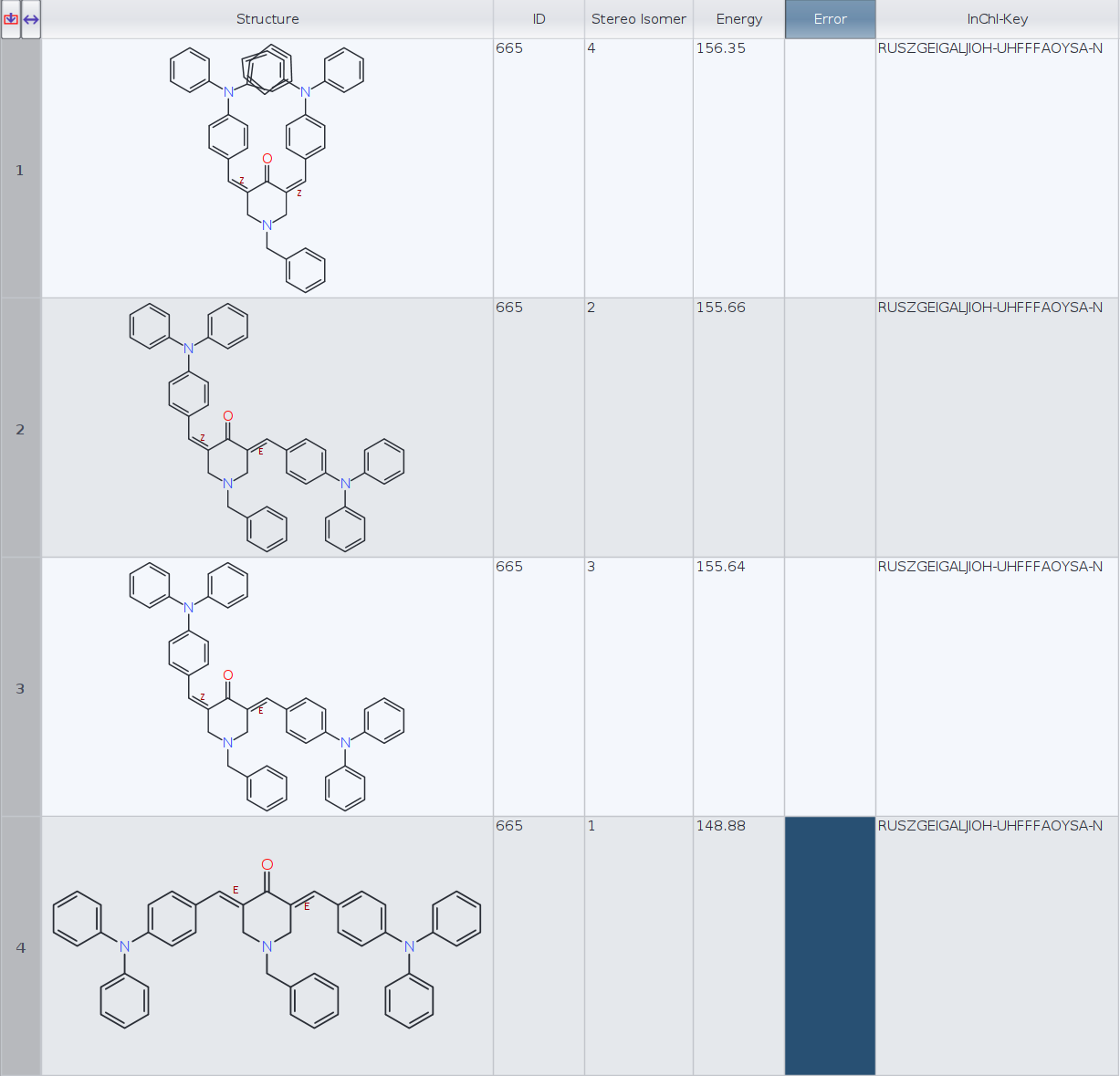
Datawarrior automatically generates the 3D geometry for four stereoisomers. Then I have generated the InChIKeys for the four molecules and Datawarrior returns the same key, RUSZGEIGALJIOH-UHFFFAOYSA-N, for every molecule (see figure).
I have tried generating the InChIKeys with RDKit in Python and instead, I have obtained the following:
ID | InChI-Key | Energy
1) RUSZGEIGALJIOH-MRSMTKAOSA-N 156.35
2) RUSZGEIGALJIOH-JYYJLMHASA-N 155.66
3) RUSZGEIGALJIOH-JYYJLMHASA-N 155.64
4) RUSZGEIGALJIOH-LMKHWIEJSA-N 148.88
I have two questions:
- Which InChIKeys are the correct ones, DataWarrior or RDKit?
- Aren't molecules 2 and 3 (E, Z and Z, E) the same stereoisomer in this particular case? RDKit InChIKeys seem to confirm that. If that is the case, perhaps DW conformer routine should only keep one of the two, or is this an intended behaviour?
Thank you for your help!
Mattia
|
|
|
|
| Re: Stereoisomers conformer generation and Inchikeys [message #1391 is a reply to message #1388] |
Thu, 02 September 2021 22:18   |
 nbehrnd
nbehrnd
Messages: 240
Registered: June 2019
|
Senior Member |
|
|
Dear Mattia,
based on visual inspection of the structure formulae, entries #2 and #3 depict the same isomer. Thus, there are three different compounds in the table for which one anticipates three different SMILES strings, InChis, or eventually InChiKeys.
Based on the visual representation, I redrew the structures in PerkinElmer's ChemDraw test page.[1] This page equally permits the export in SMILES, InChi, and InChiKey format. There is no match of the later with those you report as generated by DW. The ChemDraw InChiKeys match the ones you report by RDKit. It is possible to process ChemDraw's SMILES strings into InChiKeys by OpenBabel, too, yet with the same result; match with RDKit/ChemDraw, no match with DW.
Importing the three different ChemDraw SMILES strings into DW yields different structure representations. However already the InChi for these match each other. Since the InChikeys only are a hash of the InChi, it is not surprising that these derivates equally match each other (again). If interested, compare with attached .dwar and Emacs .org file.
Based on these observations, pending correction of this local problem for DW, I would suggest to use the InChiKeys by RDKit. On occasion, implementation of the underlying rules indeed may cause problems which remain unidentified for quite some time until comparison with other programs.[2]
Norwid
[1] https://chemdrawdirect.perkinelmer.cloud/js/sample/index.htm l#
[2] https://mattermodeling.stackexchange.com/questions/6460/rdki t-and-pysmiles-results-differ-on-some-smiles-strings
Addition:
Since DW is able to generate a random library of molecules and to assign for these SMILES and InChiKeys, I wrote a little Python script to compare InChiKeys by DW with those generated by OpenBabel. (This only is low level of concept/doodle only.) For two runs (10 and 250 molecules), the ratio of SMILES for which both programs assign different InChiKeys over the grand total of structures submitted is a little bit higher, than anticipated. Perhaps a complementary check with RDKit is useful before Thomas steps in.
[Updated on: Fri, 03 September 2021 12:27] Report message to a moderator |
|
|
|
|
|
|
|
| Re: Stereoisomers conformer generation and Inchikeys [message #1431 is a reply to message #1403] |
Sun, 31 October 2021 20:03   |
 nbehrnd
nbehrnd
Messages: 240
Registered: June 2019
|
Senior Member |
|
|
It should be noted that tautomers share standard an InChI and standard InChIKey in common.
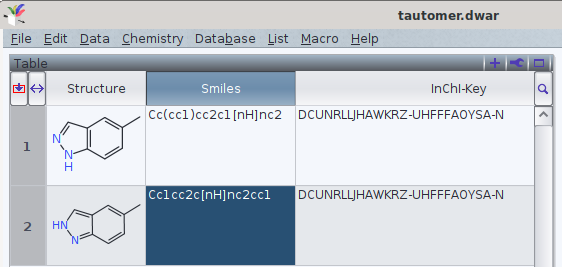
One FAQ by the InChI trust (though by 2012-05-12) explains this by
«different tautomers have the same connectivity/hydrogen layer»[1]
With the current version of InChI executables (1.06, December 2020), it is possible e.g., to add what InChI defines as «Fixed H layer», which offers to discern the tautomers. However, currently this is may yield a non-standard InChI (i.e., the string does not start with InChI=1S/, but InChI=1/). Thus, their use in a search in databases[2] may refer to more than one chemical entity.
Norwid
[1] https://www.inchi-trust.org/technical-faq-2/
[2] «Many InChIs and quite some feat» by Warr, W.A., J. Comput. Aided Mol. Des., 2015, 29: 681. https://doi.org/10.1007/s10822-015-9854-3
-
 Attachment: tautomer.dwar
Attachment: tautomer.dwar
(Size: 3.10KB, Downloaded 552 times)
-
 Attachment: tautomer.png
Attachment: tautomer.png
(Size: 23.80KB, Downloaded 1214 times)
|
|
|
|
|
|
Goto Forum:
Current Time: Mon Mar 09 11:44:31 CET 2026
Total time taken to generate the page: 0.00701 seconds
|
