| Count # scaffolds by plate ID [message #1276] |
Mon, 03 May 2021 16:35  |
 chemtv
chemtv
Messages: 28
Registered: February 2015
Location: Indianapolis
|
Junior Member |
|
|
Hi folks,
I am looking at some plated cpds where I have individual IDs and the assay plate IDs that I am trying to analyze. I want to calculate Murcko scaffolds then count the number of different scaffolds on each plate, (rough estimate of diversity). For datawarrior I suspect this is where macros can be your friend but I'm not familiar with writing macros. Has anyone else tried something like this? Even in Knime I'm not sure how to get to counts of a previous "aggregation". Any thoughts?
Thanks,
Greg
|
|
|
|
| Re: Count # scaffolds by plate ID [message #1277 is a reply to message #1276] |
Mon, 03 May 2021 18:02   |
 nbehrnd
nbehrnd
Messages: 240
Registered: June 2019
|
Senior Member |
|
|
Once the structures are read by DW, you may launch via chemistry -> analyze scaffolds the assignment of the Murcko scaffold. If you like, DW may write a new .dwar file with a frequency count of these common denominators, too:
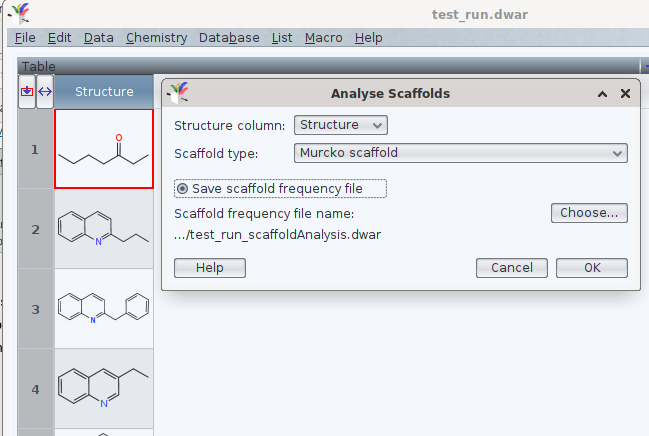
In these files, the idcode of the identified scaffold is separated by a tabulator from the integer, as documented in the archive attached below. Is this the direction of analysis you would like to automate?
If so, it suffices to identify the pathway you would go to process the data manually, and to document this by recording a macro (`macro -> start recording` to initiate, `macro -> stop recording` to complete DW's «training»). You may export then export the macro as a file as an individual .dwam file, a plain ASCII file.
The one attached below as example requires some adjustment in line #5 about the path and file name for the new .dwar (about the frequency count) to be written by you. This edit has to happen outside DW. Then, DW already working, load this macro adjusted .dwam file (macro -> import macro) and let it run (macro -> run macro, then select export_scaffold). As set up by now, it will write murcko_scaffolds.dwar as permanent record.
Note, regardless of your .dwar processed, above macro will yield a file murcko_scaffolds.dwar with the frequency count. Because of the static file name of the output it is possible you accidental overwrite the results of a precedent analysis.
Norwid
[Updated on: Mon, 03 May 2021 18:34] Report message to a moderator |
|
|
|
|
|
|
|
| Re: Count # scaffolds by plate ID [message #1280 is a reply to message #1279] |
Tue, 04 May 2021 19:06   |
 nbehrnd
nbehrnd
Messages: 240
Registered: June 2019
|
Senior Member |
|
|
I think I grasp your intent. By running the scaffold analysis, you aim for a table which either separates the entries by category like
| scafffold | frequency | plate addresses | individual label |
|-------------+-----------+-----------------+--------------------------------|
| isoxazolone | 2 | 12, 91 | cmpd18, cmpd13 |
| antipyrine | 4 | 18, 19, 20, 21 | cmpd23, cmpd24, cmpd25, cmpd26 |
| ... | ... | ... | ... |
| scafffold | frequency | plate address / individual label |
|-------------+-----------+------------------------------------------------|
| isoxazolone | 2 | 12, cmpd18; 91, cmpd13 |
| antipyrine | 4 | 18, cmpd23; 19, cmpd24; 20, cmpd25; 21, cmpd26 |
| ... | ... | ... |
Norwid
|
|
|
|
| Re: Count # scaffolds by plate ID [message #1281 is a reply to message #1280] |
Wed, 05 May 2021 06:23   |
amorrison
Messages: 39
Registered: March 2016
|
Member |
|
|
Hi Greg,
Would either of these achieve what you are aiming to do.
a) Generate Murcko scaffolds then calculate 'Frequency of same value in same category' - frequencyInCategory(category-column, value-column). Where the category-column is the plateID and the value-column is the Murcko.
b) Generate Murcko scaffolds then merge rows on both Murcko and PlateID. This would give you the list of compoundIDs with the same Murcko in one cell which you could then do a valueCount to get the number of compounds.
Best,
Angus
|
|
|
|
| Re: Count # scaffolds by plate ID [message #1282 is a reply to message #1281] |
Wed, 05 May 2021 23:23   |
 chemtv
chemtv
Messages: 28
Registered: February 2015
Location: Indianapolis
|
Junior Member |
|
|
Starting with: 500K smiles, cpdID, plateID
Here's what I'm ultimately trying to get to...
plateID | Num Scaffolds | Scaffolds (smiles of scaffold)
------------------------------------------------------------ ----------
abc01 | 5 | phenyl, pyridine, indole, thiazole, napthyl
abc02 | 3 | phenyl, pyrimidine, napthyl
abc03 | 2 | indole, oxazole
Actually the 1st two columns are what I need.
Greg
|
|
|
|
| Re: Count # scaffolds by plate ID [message #1283 is a reply to message #1282] |
Thu, 06 May 2021 14:24   |
 nbehrnd
nbehrnd
Messages: 240
Registered: June 2019
|
Senior Member |
|
|
It is possible to use the structures of one .dwar file for a comparison against structures in a second .dwar file. Because I did not identify (yet) how to point from the frequency table generated specifically to the SMILES strings of the Murcko scaffolds of the original file, I wrote a little Python script to address the initial question. The script depends on standard CPython and the RDKit library only. For a small library (say, .le. 3k entries), I perceive the rate of computation as fast enough; of course your mileage may vary when submitting 500k.
The archive, in addition to the script, equally documents a test run with said script.
Norwid
[Updated on: Thu, 06 May 2021 14:25] Report message to a moderator |
|
|
|
|
|
