| Visualising SMILES string [message #1256] |
Thu, 18 March 2021 10:55  |
amorrison
Messages: 39
Registered: March 2016
|
Member |
|
|
Hi Thomas,
I have a smiles string with an attachment point that has been created in another application, e.g. C1CCC([*:1])CC1. Datawarrior fails to identify it as a valid smiles either on paste or convert structure to name. I'm assuming it's the '*:1' that is the issue. Can I modify the smiles so it can be interpreted by Datawarrior?
many thanks in advance,
Angus
|
|
|
|
|
|
| Re: Visualising SMILES string [message #1258 is a reply to message #1257] |
Sun, 21 March 2021 05:00   |
amorrison
Messages: 39
Registered: March 2016
|
Member |
|
|
Thanks Thomas,
I've tried this. Doing a find and replace my smiles string becomes - C1CCC([*])CC1. If try a structure to name this is still not recognised as a valid smiles. If I remove the square bracket - C1CCC(*)CC1. I get the structure but lose the attachment point.
Sorry if this trivial I'm a med chemist and not so familiar with smiles syntax.
Thanks,
Angus
|
|
|
|
| Re: Visualising SMILES string [message #1261 is a reply to message #1258] |
Tue, 23 March 2021 16:38   |
 thomas
thomas
Messages: 742
Registered: June 2014
|
Senior Member |
|
|
Dear Angus,
I have extended the SmilesParser within OpenChemLib and used by DataWarrior to support '*' and '?' as pseudo atom symbols. Both symbols can be used inside or outside square brackets. While '*' is inline with the opensmiles.org standard, '?' is not. Nevertheless, because of the missing distinction in the smiles syntax between query fragments and full molecules, I believe that '?' makes sense.
'*' creates a wild card atom, which is an atom query feature and only allowed in query fragments. Therefore, a smiles containing a '*' is atomatically perceived as a query structure rather than a full molecule. Its free valences are not considered as being filled with hydrogen.
'?' is converted into an atomicNo=0, which DataWarrior uses as attachment point, e.g. in R-groups after a SAR deconvolution. These atoms are meant to not exist. Their sole purpose is to carry the bond sticking out of the R-group.
If you paste the these 6 rows of smiles:
C1CCC([*:1])CC1O
C1CCC([*])CC1O
C1CCC(*)CC1O
C1CCC([?:1])CC1O
C1CCC([?])CC1O
C1CCC(?)CC1O
into a new DataWarrior Window (newest dev release), then you get the following table:
Note that I have added an oxygen atom. If you see an 'H' at the oxygen, the structure is considered a molecule. If not, the structure is a query fragment with open valences.
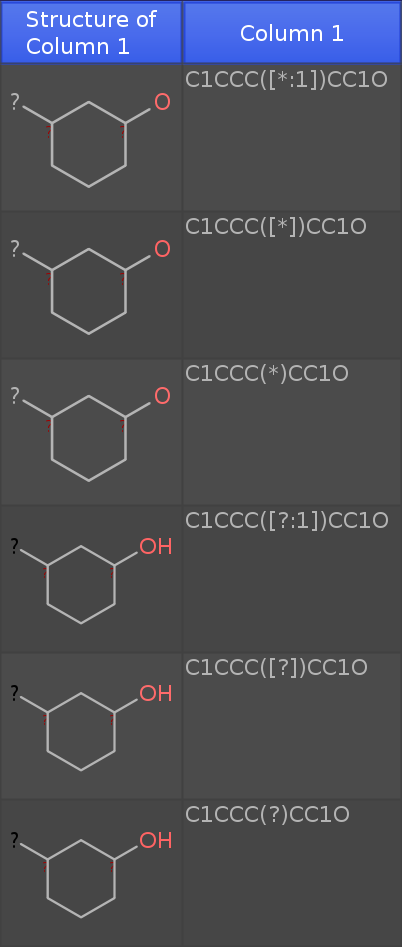
-
 Attachment: t.png
Attachment: t.png
(Size: 41.90KB, Downloaded 1416 times)
|
|
|
|
|
|
