| suggest: adjustment .sdf export [message #908] |
Fri, 15 May 2020 17:59  |
 nbehrnd
nbehrnd
Messages: 211
Registered: June 2019
|
Senior Member |
|
|
Prior to further analysis of a library,[1] its entries were deduplicated by Data ->
merge equivalent rows, using content of the structure column as sole criterion. The
work with the .sdf subsequently generated by DataWarrior worked fine if the compound
name column used the row number.
Yet, retaining the information of the molecules' name -- here, a PubChem identifier
-- may be useful as a structure may be attributed more than one.[2] The corresponding
choice of compound name column to equate automatic may then yield a .sdf which is not
understood, e.g. by openbabel (version 3.0.0, April 2020).
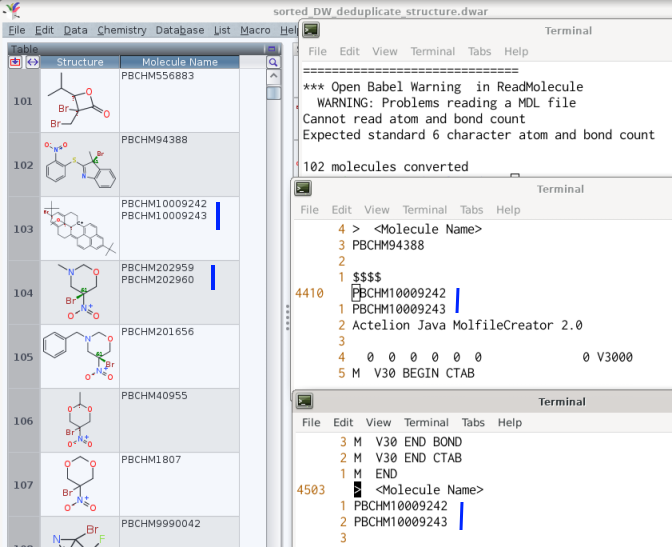
The suggestion for this type of .sdf export by DW is to report the molecules names
in the data's header / footer on one line, separated only by a blank space.
The archived .dwar equally contains cells with more then one multiple occurrence of
the same PubChem number (e.g. cell #46 about PBCHM2982, PBCHM47354, and PBCHM40585).
The desideratum for cases like this one is to retain only one occurence of each
PubChem number per cell.
[1] https://github.com/IanAWatson/Lilly-Medchem-Rules/blob/maste r/test/example_molecules.smi, revision Apr 26, 2020
[2] E.g., https://www.ncbi.nlm.nih.gov/pmc/articles/PMC4702940/
|
|
|
|
