|
|
|
|
| Re: Question about R-group occurences estimation [message #773 is a reply to message #759] |
Tue, 11 February 2020 14:53   |
 nbehrnd
nbehrnd
Messages: 224
Registered: June 2019
|
Senior Member |
|
|
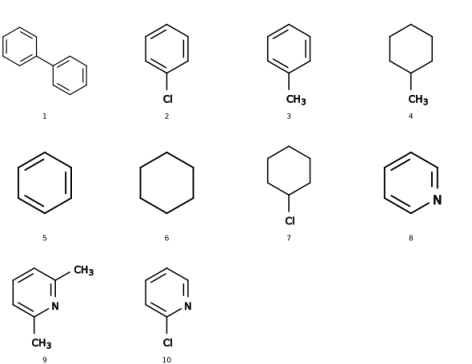
There are two possible difficulties to retrieve methyl groups as-such in a list of SMILES. For one, the characteristic of them is the (single) carbon atom and searching just for this is less identifying than the string of c1ccccc1 about benzene, for example. Second, probably there would be a need to add explicit hydrogens on all SMILES before these would be easier to identify (which may be done, e.g., with babel).
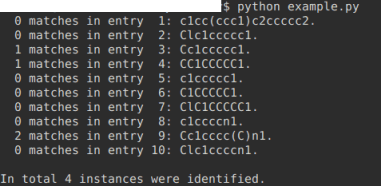
If you have access to Python, then the additional module by rdkit (http://rdkit.org/) may be quite helpful to querry your SMILES with SMARTS. With the test file of smiles_list.smi attached below, the identification of methyl groups (in SMART's convention, expressed as [CH3]) works fine both locally -- per SMILES entry -- as well as in counting the globally:
from rdkit import Chem
smiles_source = "smiles_list.smi"
grand_total = 0
# example pattern to identify and count:
functional_group = Chem.MolFromSmarts('[CH3]') # methyl group
# alternative examples:
#functional_group = Chem.MolFromSmarts('c1ccccn1') # for a pyridine
#functional_group = Chem.MolFromSmarts('C1CCCCC1') # for cyclohexane
with open(smiles_source, mode="r") as source_file:
for index, line in enumerate(source_file, start=1):
molecule = Chem.MolFromSmiles(line.strip())
match = molecule.GetSubstructMatches(functional_group)
print("{:3} matches in entry {:2}: {}.".format(
len(match), index, line.strip()))
grand_total += len(match)
print("\nIn total {} instances were identified.".format(grand_total))
As DataWarrior relies on java, the implementation of the «ErtlFunctionalGroupsFinder» described by Fritsch et al. ( https://jcheminf.biomedcentral.com/articles/10.1186/s13321-0 19-0361-8, open access) equally may be of interest for Thomas.
-
 Attachment: example.png
Attachment: example.png
(Size: 30.77KB, Downloaded 938 times)
-
 Attachment: listing.png
Attachment: listing.png
(Size: 51.00KB, Downloaded 821 times)
-
 Attachment: smiles_list.smi
Attachment: smiles_list.smi
(Size: 0.12KB, Downloaded 409 times)
-
 Attachment: example.py
Attachment: example.py
(Size: 0.76KB, Downloaded 398 times)
|
|
|
|
| Re: Question about R-group occurences estimation [message #775 is a reply to message #773] |
Wed, 12 February 2020 19:40  |
 thomas
thomas
Messages: 711
Registered: June 2014
|
Senior Member |
|
|
This is now built in: "Chemistry->From Chemical Structure->Add Substructure Count..."
This opens a dialog to
-select the structure column
-to draw a fragment with optional query features
-and to define whether counted fragments may overlap on some atoms
It is in the current beta and will be in the next official version that is due in a few days
|
|
|
|
