Home » DataWarrior » Functionality » feature suggest: array cleaning by delete button
| feature suggest: array cleaning by delete button [message #2024] |
Fri, 27 October 2023 12:33  |
 nbehrnd
nbehrnd
Messages: 211
Registered: June 2019
|
Senior Member |
|
|
Dear Thomas,
I would like to suggest an extension to edit the structure array (presented after the generation of an evolutionary library); to delete selected entries by single press of the delete key.
While browsing an EL (attached), I noticed a few entries either with a `&1` as place holder for (R)/(S) on stereochemical centres, or «crossed double bonds» to indicate an unassigned/flexible (E)/(Z) configuration. For small test down the road, my intent was to select these records (left mouse click) -- eventually multiple (left mouse click while holding the Ctrl key) -- and to remove them.
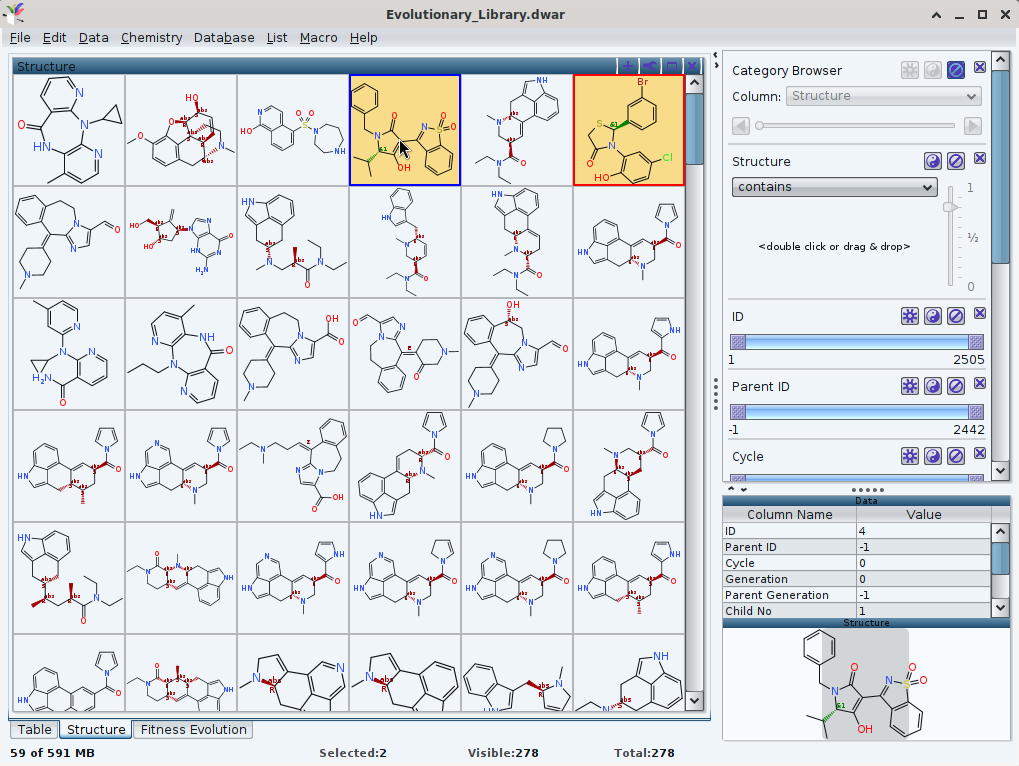
At present, this is possible by the subsequent Data -> Delete Rows [sic!] -> Selected Rows. This approach works to remove only the selected entries (thankfully: only the ones identified, and not the whole row of the entries). My suggestion is to offer a remove of the entries already marked by single press of the delete button as a shorter one.
The pulldown menu available for the array (right mouse button click on a cell) includes an entry «edit», but no delete. If the suggested functionality were provided here, it would again request a journey of three mouse clicks instead of a single key action. To change to the tab «table», to select whole rows then deleted is possible though this display does not offer as much a synopsis of the structures in question as does the tab «structures».
The observation refers to DW for Linux in version 05.06.00 by September 22nd.
Regards,
Norwid
|
|
|
|
| Re: feature suggest: array cleaning by delete button [message #2025 is a reply to message #2024] |
Thu, 02 November 2023 12:00   |
 thomas
thomas
Messages: 655
Registered: June 2014
|
Senior Member |
|
|
Dear Norvid,
many thanks for the suggestion. My personal feeling would be that it would be dangerous, if one could directly delete rows from the data with one key, especially since there is no UNDO functionality. However, currently there is already a way to remove individual rows one by one with one key-stroke using a row list:
- select one or more rows, e.g. in the structure view
- press CTRL-1, which adds all selected rows to a default row list and creates a filter for that.
- select 'Default List' in the filter and invert the filter. Now all molecules in the default list are invisible
- select more structures and press CTRL-1 to add them to the list, which is to hide the from the view
- Choose "Data->Delete Rows->Invisible Rows"
(before deleting them, you may invert the filter again to see and check all molecules about to be deleted)
Would this be a viable alternative to the direct deletion key?
PS: &1 is the indicator for a racemic stereo center, where the '1' is the groups number in case we have multiple stereo centers that have relative stereo configurations, but are racemic none the less
Best wishes,
Thomas
|
|
|
|
| Re: feature suggest: array cleaning by delete button [message #2026 is a reply to message #2025] |
Sat, 04 November 2023 20:19  |
 nbehrnd
nbehrnd
Messages: 211
Registered: June 2019
|
Senior Member |
|
|
Dear Thomas,
thank you for depicting a different logic to address the task. Beside the possibility to go back and forth between the original set of data and the entries to be removed, it offers to export the unwanted entries (and their data associated) for a later review -- and hence, an additional advantage to the «single click» approach suggested.
There then however is a (cosmetic?) issue. After the removal of the unwanted structures, the counter of selected entries reverts to zero, the counter of the total of structures decreased. So far, I assumed this integer equally should be the one shown as "structures visible", quite like the state prior to edit/removal of individual structures (even before their identification and selection).
The EL in question was generated with the default set of 8 motifs (two of them about (R)/(S) racemates, i.e. a total of 10 molecules), the creation of compounds like approved drugs with a sole fitness criterion (molecular weight equal or less equal than 400). This lead to 278 records, where 2 records contain one single stereogenic centre each which might be either (R), or (S). Plus 5 other records with a C=C double bond in either (E), or (Z) configuration.* This would lower the total count either by 7 (for a then total equal to 271), or by 14 (to then retain 264 structures) if one aims for a representation without either (E)/(Z), nor (R)/(S) flexibility. Application of the Data -> Delete Rows -> Invisible Rows however yields a "Selected: 0, Visible: 237, Total: 271". (Simultaneously to the edit applied, the slider "generation" turns red, too.).
The export of the results as .txt file contains 271 records. Saving the (edited) .dwar file as a new .dwar file, and subsequent reading of this new .dwar file still displays the reading "selected: 0, visible: 237, total 271" on the lower rim of the frame, too. Is there a discrepancy between the selection of individual entries of a row displayed vs eventual edit (i.e. the remove) of of every entry of their corresponding rows displayed? My intent was the remove of only the 7 potentially ambiguous entries.
Best regards,
Norwid
\* Somehow the submission of this original EL .dwar file to DW's Generate Conformers with a constrain of maximal 1 conformer per stereo isomer is able to split/disentangle them, i.e. to provide one (R) and one (S) conformer; or one (E) and one (Z) conformer (DW for Linux, v05.06.00, 22-Sep-2023).
The centre's assignment by `&` and `or` in the array display, where applicable, is the same throughout the program, hence known.
|
|
|
|
Goto Forum:
Current Time: Sat Apr 27 23:18:03 CEST 2024
Total time taken to generate the page: 0.03167 seconds
|
