| Re: Molecule style & color [message #990 is a reply to message #989] |
Thu, 09 July 2020 09:57   |
 nbehrnd
nbehrnd
Messages: 224
Registered: June 2019
|
Senior Member |
|
|
Hello Paul,
a quick work-around could employ the platform independent Jmol.(1) It is possible to
save all of the conformers generated in one .sdf file in common -- just take care that
the export retains the 3D coordinates (which is not the default).
In Jmol running, the entry File -> Console opens the programs' own little terminal.
Then indicate Jmol the .sdf to read; it will recognize that the .sdf contains multiple
models, but by default will display only the first one. This is the chance to disable
the display of double / aromatic / triple bonds with more than one line (by the second
command), to adjust the diameter of the sticks and balls (third command) and eventually
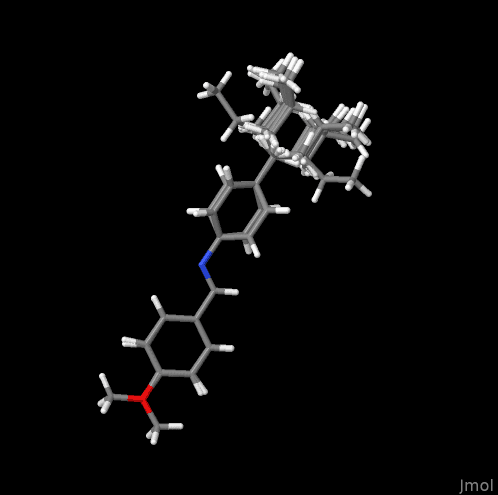
display all models (here: all superimposed conformers) at once by the instruction of
model 0.
The scene may be exported like below.
The interaction with Jmol may be scripted (e.g., 2,3,4) thus you may reuse instructions
more than once. If the color of an atom type does not fit your needs, for example, you
could alter the representation by
select hydrogen
color gray
for all models load at once. You may return to the default color scheme by
select all
color cpk
Norwid
1) http://wiki.jmol.org/index.php/Main_Page
2) https://chemapps.stolaf.edu/jmol/docs/
3) https://earth.callutheran.edu/Academic_Programs/Departments/ BioDev/omm/jsmol/scripting/molmast.htm#V
4) Hanson, J.Appl.Cryst.(2010).43, 12501260, doi: 10.1107/S0021889810030256
|
|
|
|
